Investigation of denaturation processes induced by guanidine hydrochloride (GdnHCl) of proteins possessing βbarrel structure porcine odorantbinding protein (OBP) and a number of fluorescent proteins (FPs) Fluorescent proteins represent a large family of proteins with unique spectroscopic properties, ie theTo the different denaturation mechanism of alphaamylase with the two denaturing agents The numbers of the refolded intermediates of ureaunfolded alphaamylase were found to be more than that denatured by GuHCl, because GuHCl may make the changes in the surface of alphaamylase molecules, by contrast, urea may do the1/11/1998 · The Sequential Mechanism of Guanidine HydrochlorideInduced Denaturation of cAMP Receptor Protein from Escherichia coli A Fluorescent Study Using 8Anilino1Naphthalenesulfonic Acid Jedrzej Malecki 1 &

Kinetic Evidence For A Two Stage Mechanism Of Protein Denaturation By Guanidinium Chloride Pnas
How does guanidine hydrochloride denature proteins
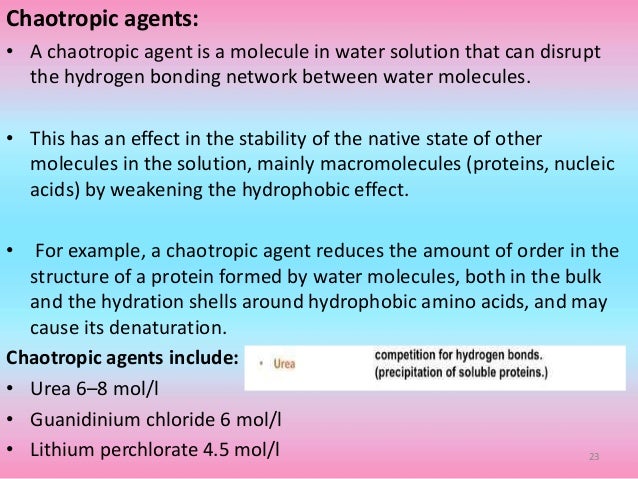
How does guanidine hydrochloride denature proteins-24/2/09 · The mechanism by which urea and guanidinium destabilize protein structure is controversial We tested the possibility that these denaturants form hydrogen bonds with peptide groups by measuring their ability to block acid and basecatalyzed peptide hydrogen exchangeGuanidine is the compound with the formula HNC(NH 2) 2It is a colourless solid that dissolves in polar solvents It is a strong base that is used in the production of plastics and explosivesIt is found in urine as a normal product of protein metabolismA guanidine moiety also appears in larger organic molecules, including on the side chain of arginine



Comparative Refolding Of Guanidinium Hydrochloride Denatured Bovine Serum Albumin Assisted By Cationic And Anionic Surfactants Via Artificial Chaperone Protocol Biophysical Insight Sciencedirect
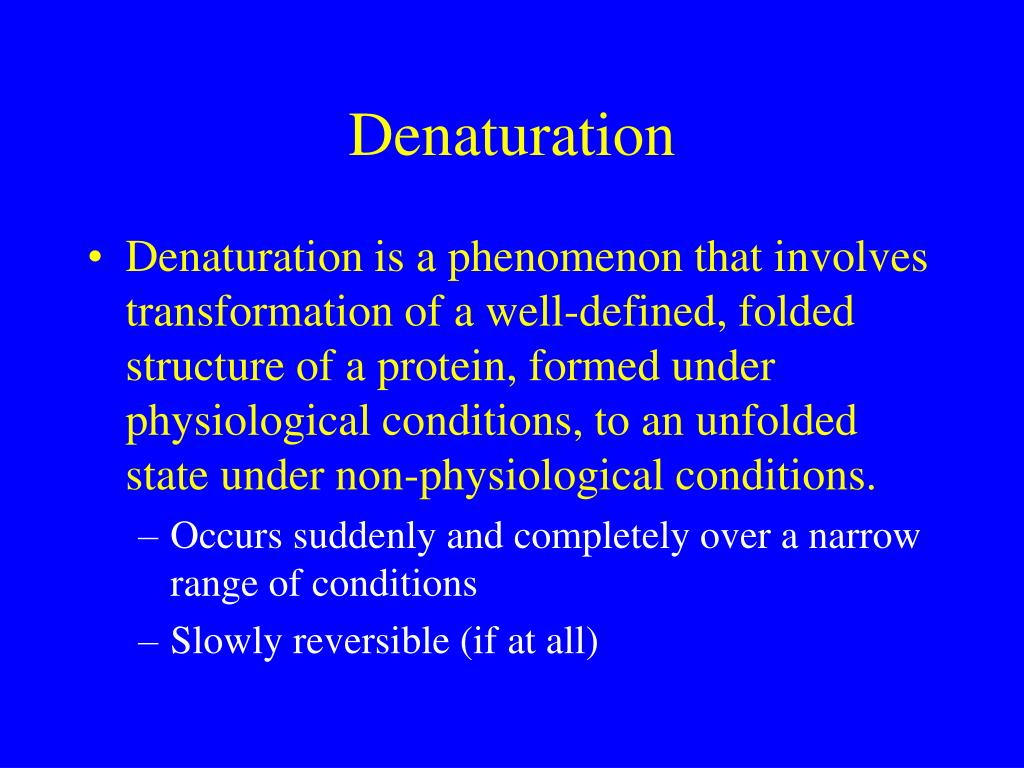
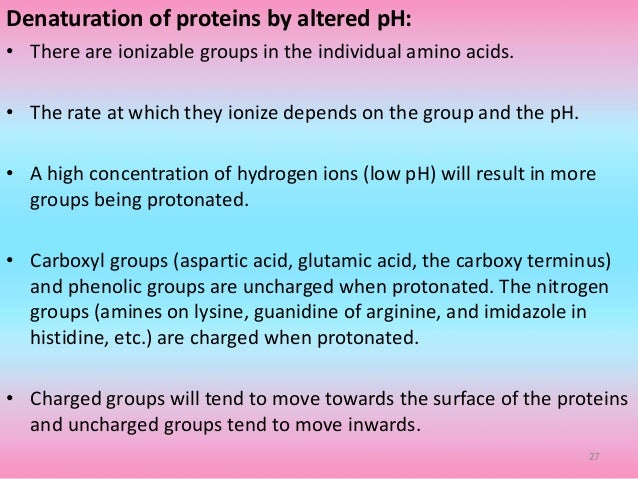
In general, as commonly used reagents in protein denaturation, guanidine hydrochloride () has relatively strong dissolving capacity and denatured ability and does not cause the covalent modification of the recombinant protein, but has the disadvantages of high cost, easy precipitation under acidic conditions, and interference with protein ion exchange chromatography;Guanidine Hydrochloride is the hydrochloride salt form of guanidine, a strong basic compound with parasympathomimetic activityGuanidine hydrochloride enhances the release of acetylcholine following a nerve impulse and potentiates acetylcholine actions on muscarinic and nicotinic receptors It also appears to slow the rates of depolarization and repolarization of muscle cell6 Second, unfolding is more likely to approach a twostate mechanism And, third, denaturation is more likely to be completely re versible
Investigation of the Mechanism of Protein Denaturation by Guanidine HydrochlorideInduced Dissociation of InhibitorProtease In this communication we describe an approach in which guanidine hydrochlorideinduced dissociation of a protein inhibitorserine protease complex is used to explore the molecular basis of protein denaturationSummary This chapter contains sections titled Historical Perspective How Urea Denatures Proteins Linear Extrapolation Method ΔG(H2O) m‐Values Concluding Remarks Experimental ProtocolsIrreversible thermal denaturation Here we report the determination of thermodynamic parameters of unfolding (H, G, and C p) for FGF1 using differential scanning calorimetry (DSC) The thermal denaturation is demonstrated to be twostate and reversible upon the addition of low concentrations of added guanidine hydrochloride (GuHCl)
Protein denaturation with guanidine hydrochloride or urea provides a different estimate of stability depending on the contributions of electrostatic interactions Oscar D Monera Department of Biochemistry and the Protein Engineering Network of Centres of Excellence, University of Alberta, Edmonton, Alberta T6G 2H7, CanadaThe protein greatly loses its structure at 6 M urea and at 8 M it is a random coil The urea induced denaturation follows twostate rule in which Native>Denatured state transition occurs in a single step whereas in case of GdnHCl, intermediates or nonnative states are observed at lower concentrations of denaturantSpectroscopy (3) Similarly, guanidine hydrochloride (GdnHCl) denaturation of serum albumin has been reported to follow both singlestep and twostep transitions (4, 10) Different probes used in denaturation studies either require a sophisticated costly instrument or higher protein concentration to study protein denaturation


D Protein Folding Stability In Vivo And In Vitro


Differences In The Pathways Of Proteins Unfolding Induced By Urea And Guanidine Hydrochloride Molten Globule State And Aggregates
Denaturation of Bovine Carbonic Anhydrase B by Guanidine Hydrochloride A PROCESS INVOLVING SEPARABLE SEQUENTIAL CONFORMATIONAL TRANSITIONS* (Received for publication, June 1, 1973) KINPING WONG$ AND CHARLES TANFORD$ From the Department of Biochemistry, Duke University Medical Center, Durham, North CarolinaFluorescence and circular dichroism spectroscopy as well as analytical ultracentrifugation and glutaraldehyde crosslinking were utilized to evaluate the tertiary and quaternary structural changes occurring on the denaturation and reconstitution pathways of transthyretin (TTR) as a function of guanidine hydrochloride (GdnHCl) concentrationInvestigation of the Mechanism of Protein Denaturation by Guanidine Hydrochloride YouTube Web http//wwweurekaselectcom/1057/articleTitle Investigation of the Mechanism of Protein


1 Outcome 2 Keeping Proteins In Their Native States Ppt Download


Denaturation Of Proteins
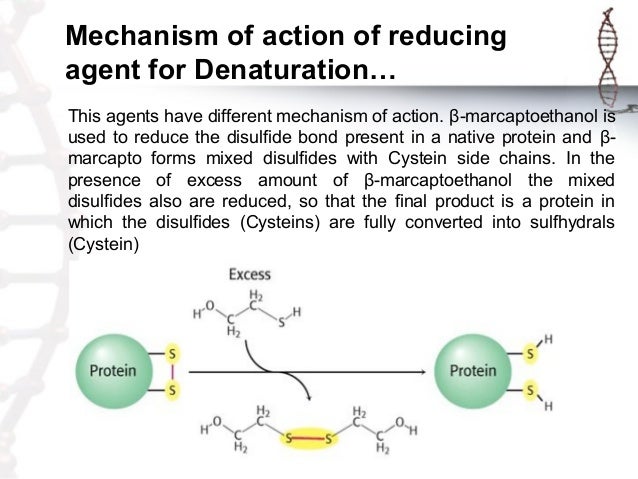
Guanidine hydrochlorideinduced denaturation of the colicin E1 channel peptide The results of unfolding analysis showed that the channel peptide's unfolding mechanism involves an intermediate structure stabilized by the Cterminal hydrophobic core of the peptide Protein Denaturation Protein FoldingInfluence of urea and guanidine hydrochloride on lysozyme stability and thermal denaturation;Denaturation of human copperzinc superoxide dismutase by guanidine hydrochloride a dynamic fluorescence study Biochemistry, 1992 Enrico Gratton Download PDF Download Full PDF Package This paper A short summary of this paper 37



Adapting The Chemical Unfolding Assay For High Throughput Protein Screening Using Experimental And Spectroscopic Corrections Sciencedirect


Destruction And Building Of The Secondary And Tertiary
30/11/10 · Protein unfolding induced by chemical denaturants such as urea and guanidine hydrochloride (GdnHCl) is a common approach to study protein folding in vitro Meanwhile, it has been shown that low concentrations of GdnHCl can cause protein stabilization by eliminating the strains in protein caused by the electrostatic interactions of charged groups on its surface 2 ,A correlation between activity, protein dynamics and conformational changes Physical Chemistry Chemical Physics, 10Kinetics of Actin Unfolding Induced by Guanidine Hydrochloride as a result of heat denaturation (2941), moderate urea or GdnHCl1 concentration (30, 38), To elucidate the mechanism of inactivated actin formation, the kinetics of GdnHClinduced actin


Add Your Page Title



The Rough Energy Landscape Of Superfolder Gfp Is Linked To The Chromophore Abstract Europe Pmc
1/1/1986 · First, the product is better defined because the degree of unfolding is maximized Proteins in 8 M urea or 6 M GdnHCI with their disulfide bonds broken approach a randomly coiled conformation?Guanidine hydrochloride (GmHCl) as a protein denaturant was first noted by Greenstein in 1938 (11) At present, urea and GmHClr are the most frequently used protein denaturants The main advantage of these denaturants is that the extent of un folding is generally greater than can be achieved by other meansObserved in the chemical denaturation of lysozyme by guanidine hydrochloride Moreover, it was found that the Raman bands of the groups on the surface of lysozyme changed before those of the other groups This indicates that the chemical denaturants interact with the protein surface before the protein core in each step and



Quantitative Calorimetric Evidences Into Counteraction Mechanism Of Denaturing Effect Of Guanidine Hydrochloride By Citrulline And Betaine Sciencedirect



Urea A And Guanidinium Hydrochloride Gdnhcl B Induced Unfolding Download Scientific Diagram
Surprisingly, however, the molecular mechanism of its action is still poorly understood Here, we provide direct kinetic evidence for the hypothesis that GdmCl unfolds proteins by a twostep mechanism In the first step, it binds to the protein surface, resulting in8/6/19 · The reversibility of the binding of human apolipoprotein AI (apo AI) to phospholipid has been monitored through the influence of guanidine hydrochloride (GdnHCl) on the isothermal denaturation and renaturation of apo A1/dimyristoylphosphatidylcholine (DMPC) complexes at 24 degree C Denaturation was studied by incubating discoidal 1100 and vesicular 1500 mol/molHome / Protein and Peptide Letters, Volume , Number 2 Investigation of the Mechanism of Protein Denaturation by Guanidine HydrochlorideInduced Dissociation of InhibitorProtease Complexes Buy Article


Protein



Urea And Guanidinium Chloride Denature Protein L In Different Ways In Molecular Dynamics Simulations Abstract Europe Pmc
The competitive process among CK, the osmolytes and GdmCl indicated that protein–solvent interactions also played a key role in the osmolyte protection of CK against guanidine denaturation Commonly occurring organic osmolytes are strongly excluded from the ordered water that surrounds proteins and therefore stabilize the protein structures 10 , 40 , 55 , 5622/1/09 · Cystatins essentially regulate lysosomal cysteine protease besides affecting several physiological processes In the present study, denaturation of a high molecular weight cystatin (Mr 664 kDa) purified from goat lung (GLCI) has been studied by monitoring its inhibitory activity, intrinsic fluorescence, circular dichroism (CD), and binding of ANSIrreversible thermal denaturation Here we report the determination of thermodynamic parameters of unfolding (DH, DG, and DC p) for FGF1 using differential scanning calorimetry (DSC) The thermal denaturation is demonstrated to be twostate and reversible upon the addition of low concentrations of added guanidine hydrochloride (GuHCl)



The Stability Of A Three State Unfolding Protein Intechopen



Urea And Guanidinium Chloride Denature Protein L In Different Ways In Molecular Dynamics Simulations Biophysical Journal
Read "The Sequential Mechanism of Guanidine HydrochlorideInduced Denaturation of cAMP Receptor Protein from Escherichia coli A Fluorescent Study Using 8Anilino1Naphthalenesulfonic Acid, The Protein Journal" on DeepDyve, the largest online rental service for scholarly research with thousands of academic publications available at your fingertipsProtocol for Guanidinium Denaturation Studies I Make Stock Solutions A Make ~ 75 M Gua·HCl titration 1 Dissolve 358 g Gua·HCl in ~30 mL water 2 Volume will expand 3 Add small amounts of water until solid is entirely dissolved 4 Bring volume up to 500 mL 5 Measure concentration of solution by refractive index measurements (Pace CN, Meth15/6/08 · To understand the different mechanisms that trigger denaturation of Protein L in urea and GdmCl, we performed a set of ns simulations at 300 K, at which temperature the protein is not able to escape from its native basin (the RMSD is lower than 3 Å) and consequently can provide structural information about the native metastable state



Guanidine Hcl Induced Denaturation Only The Transition Curves Of Download Scientific Diagram



Cooperative Protein Unfolding A Statistical Mechanical Model For The Action Of Denaturants Sciencedirect
Aminomethanamidine hydrochloride guanidine chlorhydr ate Guanidine HCl is a normal product of protein metabolism and a protein denaturant;Target Guanidine HCl is the most popular protein denaturant Analysis of unfolding transitions by Guanidine HCl provides several important parameters regarding the mechanism of conformationalOur results agree with the general consensus that the denaturing effect of guanidine hydrochloride is due to its favorable interaction with the polar parts of proteins and that the nonpolar side chains have no or little favorable interaction with guanidine hydrochloride2Protein Science Laboratory of the Ministry of Education, School of Life Science and Engineering, Tsinghua University, Beijing, China


Ppt Protein Denaturation Powerpoint Presentation Free Download Id


Probing Conformational Stability And Dynamics Of Erythroid And Nonerythroid Spectrin Effects Of Urea And Guanidine Hydrochloride
Guanadine HCL is one of the strongest denaturing agents for protein folding studies, and is used to denature proteins due to its chaotropic qualities21/2/18 · Protein denaturation, mechanism, causes, • Certain reagents such as urea and guanidine hydrochloride denature proteins by forming hydrogen bonds to the protein groups that are stronger than the hydrogen bonds formed between the groupsProtein Denaturation Urea vs Guanidine Hydrochloride (GdnHCl) In equilibrium denaturation experiments, the concentration of urea at midpoint of denaturation is always higher than that of
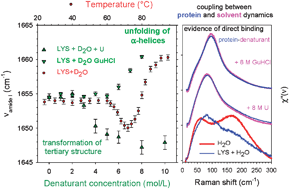


Influence Of Urea And Guanidine Hydrochloride On Lysozyme Stability And Thermal Denaturation A Correlation Between Activity Protein Dynamics And Conformational Changes Physical Chemistry Chemical Physics Rsc Publishing



Enzymatic Degradation Of Rna Causes Widespread Protein Aggregation In Cell And Tissue Lysates Embo Reports
Guanidine hydrochloride (GdnHCl) give the same estimates of the stability of a particular protein Moreover, estimates of protein stability from GdnHCl and urea denaturation data might differGuanidine hydrochloride (GdnHCl)induced unfolding of bovine spleen galectin1 (Gal1) exhibits threestate mechanism involving exclusive, structured tertiary monomer in 05 M GdnHCl Gal1 has one tryptophan residue (Trp 68) per subunit15/2/11 · To evaluate the role of the hydration layer on the protein surface of actomyosin, we compared the effects of urea and guanidineHCl on the sliding velocities and ATPase activities of the actinheavy meromyosin (HMM) system Both chemicals denature proteins, but only urea perturbs the hydration layer



Guanidine Hydrochloride Denaturation Of Sc S100a4 Proteins A Download Scientific Diagram


Protein Denaturation
In order to clarify the mechanism of denaturantinduced unfolding of proteins we have calculated the interactions between hydrophobic and ionic species in aqueous guanidinium chloride and urea solutions using molecular dynamics simulations Hydrophobic association is not significantly changed in urea or guanidinium chloride solutionsMolecular mechanism for osmolyte protection of creatine kinase against guanidine denaturation WenBin Ou1, YongDoo Park1 and HaiMeng Zhou1,2 1Department of Biological Sciences and Biotechnology, Tsinghua University, Beijing, China;Tration of urea or guanidine hydrochloride which weaken both hydrophobic interactions and hydrogen bonds in the protein mole cule1†`3) These studies, in connection with certain physical data, indicated that hydrogen bonding in the secondary structure of en zymes was essential to maintain their cata



Urea And Guanidinium Chloride Denature Protein L In Different Ways In Molecular Dynamics Simulations Biophysical Journal



Figure 7 From Interactions Between Hydrophobic And Ionic Solutes In Aqueous Guanidinium Chloride And Urea Solutions Lessons For Protein Denaturation Mechanism Semantic Scholar
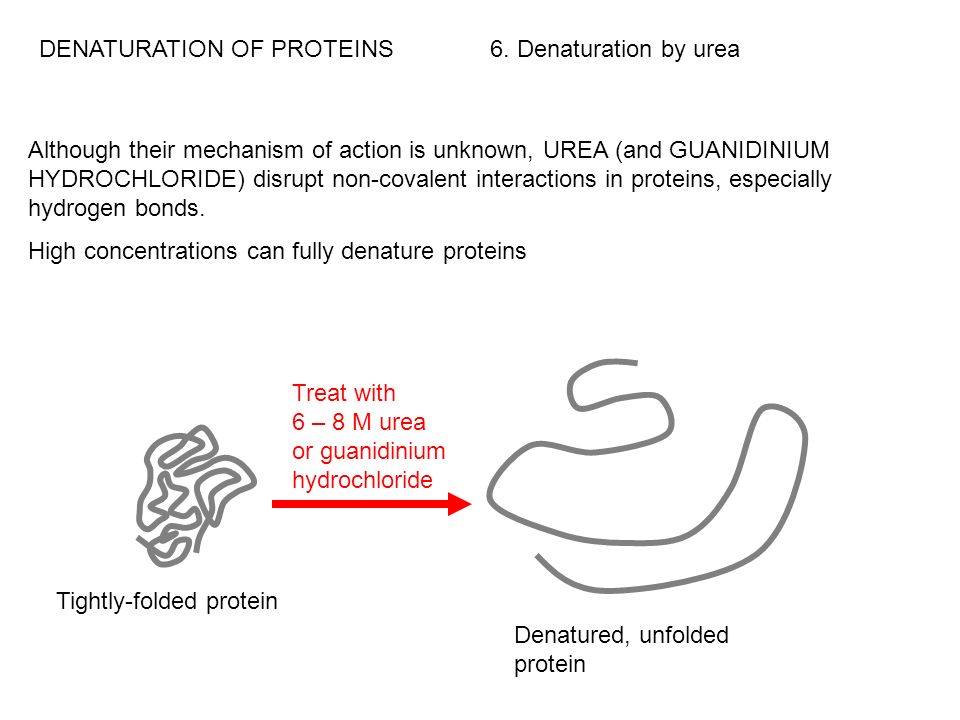
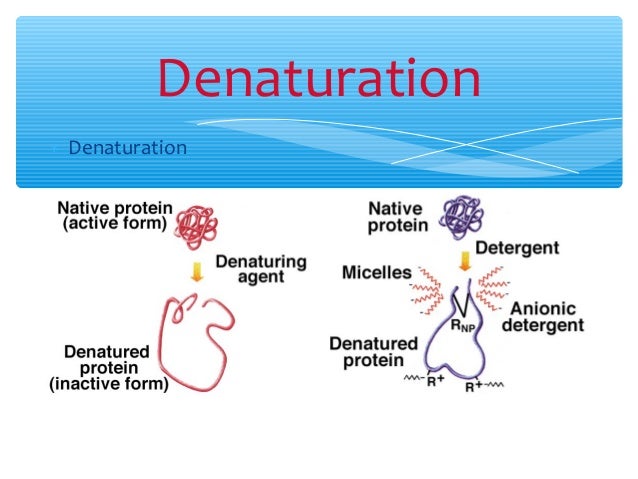
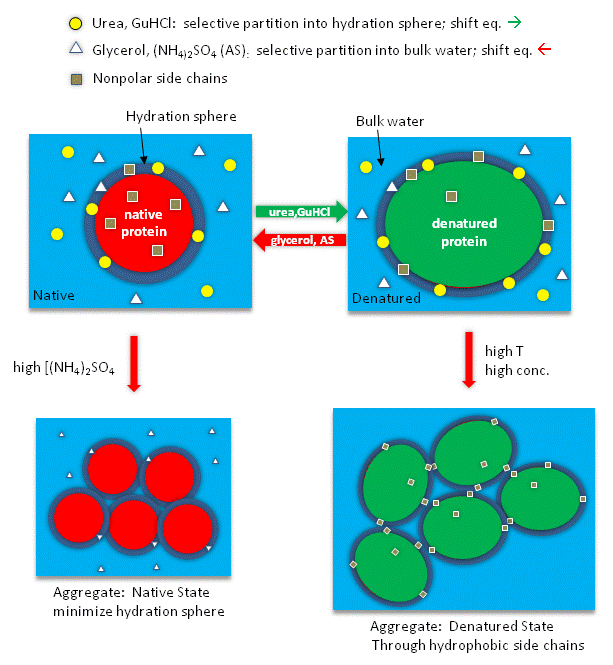
Guanidine hydrochloride (GdnHCl) is hydroscopic and a small molecule It is a potent denaturant of enzymes, which results in conformational changes in the enzyme, leading to complete loss of enzyme activity GdnHCl is involved in blocking the activity of heat shock protein 104 (Hsp104) adenosine triphosphatase (ATPase) in vivo1/4/14 · Guanidinium chloride (GdmCl) has been used to modulate the stability of proteins for more than 50 years;22/3/21 · 1Guanidine hydrochloride can denature proteins Proteins are easily affected by external factors (temperature, denaturant, etc) and undergo conformational changes, loss of activity, abnormal changes in physicochemical properties, a process known as protein denaturation Guanidine hydrochloride is an important denaturant that produces strong denaturation effects, including two denaturation mechanisms



Characterization Of The Nucleoside Triphosphatase Activity Of Poliovirus Protein 2c Reveals A Mechanism By Which Guanidine Inhibits Poliovirus Replication Journal Of Biological Chemistry
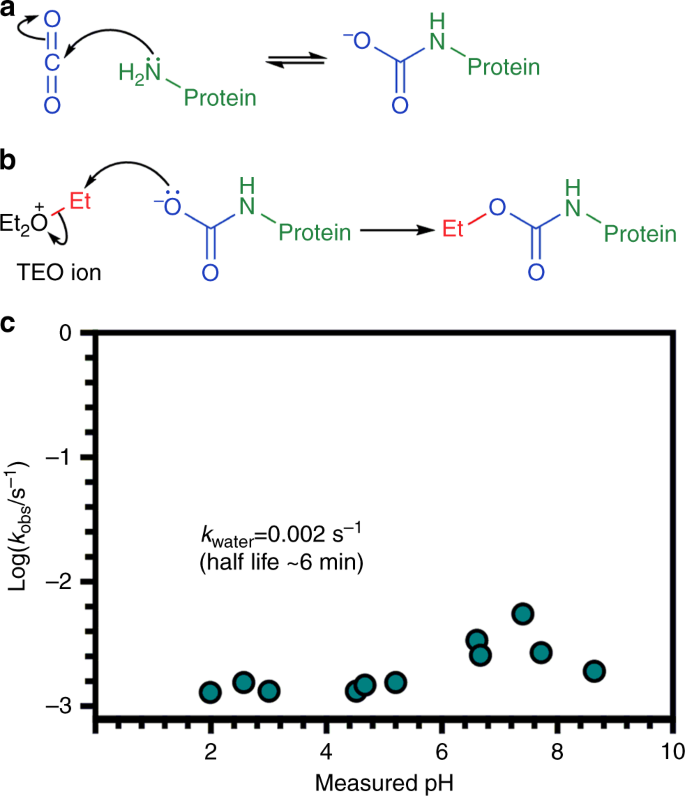


The Identification Of Carbon Dioxide Mediated Protein Post Translational Modifications Nature Communications
Firstly, the mechanism of protein denaturation of guanidine hydrochloride and urea were illustrated There are three suggested mechanisms for the above denaturants The first statement is the hydrogen bond broken which make the protein structure flexible The second one is the denaturant which deteriorate the hydrophobic effect



Comparative Refolding Of Guanidinium Hydrochloride Denatured Bovine Serum Albumin Assisted By Cationic And Anionic Surfactants Via Artificial Chaperone Protocol Biophysical Insight Sciencedirect



Kinetic Evidence For A Two Stage Mechanism Of Protein Denaturation By Guanidinium Chloride Pnas



The Peptide Bond Protein Structure Protein Denaturation Properties



Protein Denaturation With Guanidinium A 2d Ir Study Topic Of Research Paper In Chemical Sciences Download Scholarly Article Pdf And Read For Free On Cyberleninka Open Science Hub



Effects Of Urea A And Guanidine Hcl B On The Activity Of Download Scientific Diagram



Ph Corrections And Protein Ionization In Water Guanidinium Chloride Biophysical Journal
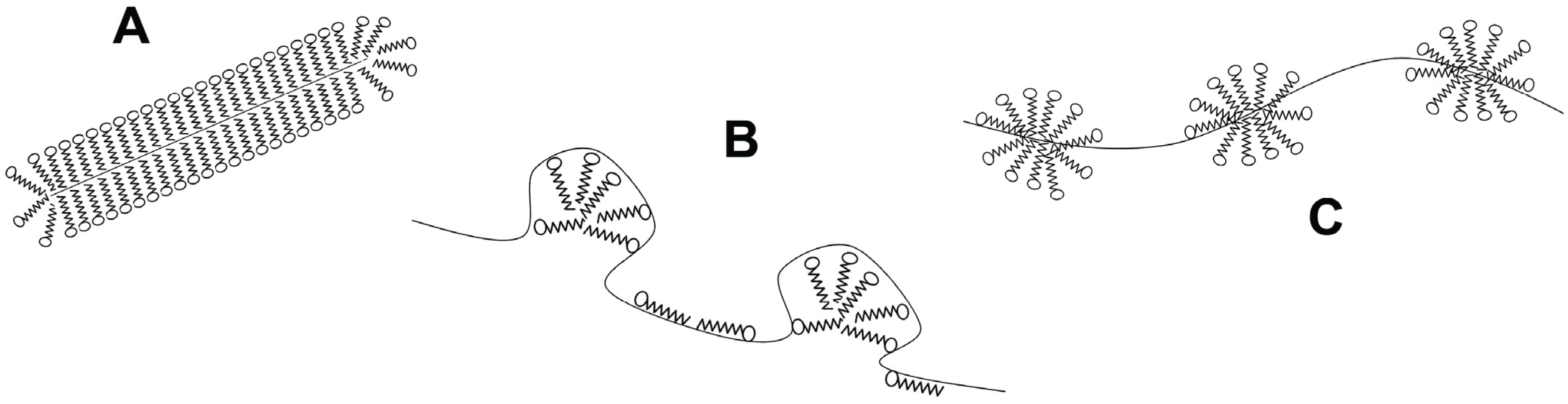


Biomolecules Free Full Text Kinetics And Thermodynamics Of Membrane Protein Folding Html



Pdf Kinetic Evidence For A Two Stage Mechanism Of Protein Denaturation By Guanidinium Chloride Semantic Scholar
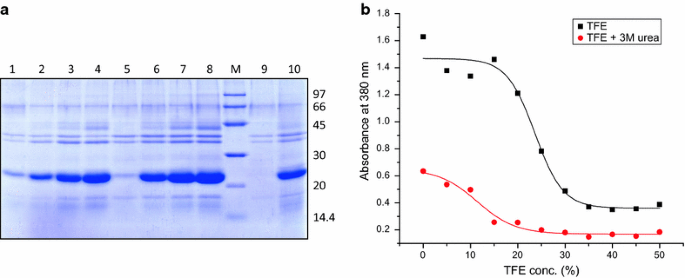


Recovery Of Bioactive Protein From Bacterial Inclusion Bodies Using Trifluoroethanol As Solubilization Agent Microbial Cell Factories Full Text
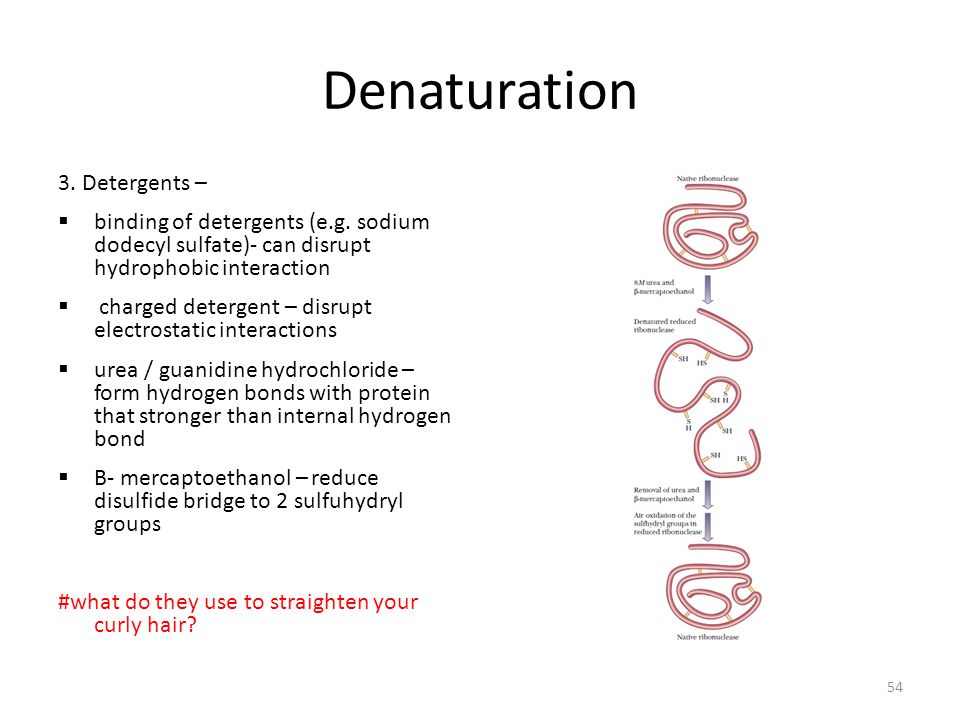


Topic 2 Biomolecule 1 Dna Protein Ppt Video Online Download
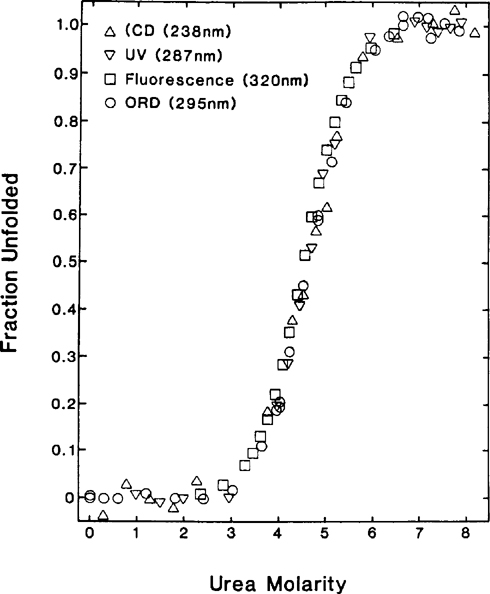


Urea And Guanidine Hydrochloride Denaturation Curves Springerlink



Investigation Of The Mechanism Of Protein Denaturation By Guanidine Hydrochloride Youtube
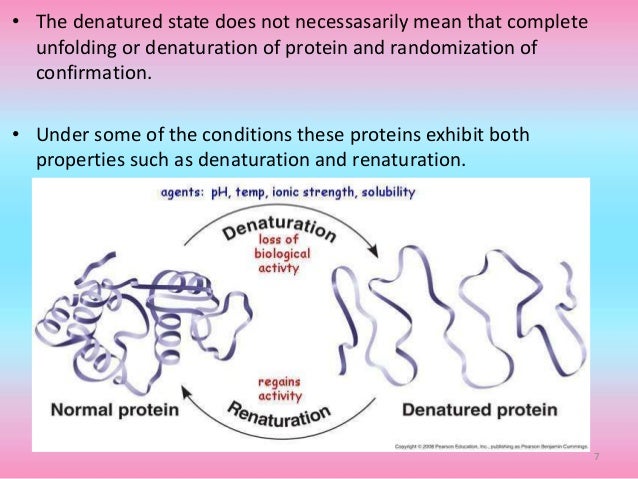


Protein Denaturation



Kinetics And Thermodynamics Of Membrane Protein Folding Topic Of Research Paper In Biological Sciences Download Scholarly Article Pdf And Read For Free On Cyberleninka Open Science Hub



Protein Stiffening And Entropic Stabilization In The Subdenaturing Limit Of Guanidine Hydrochloride Biophysical Journal



Figure 4 From Interactions Between Hydrophobic And Ionic Solutes In Aqueous Guanidinium Chloride And Urea Solutions Lessons For Protein Denaturation Mechanism Semantic Scholar
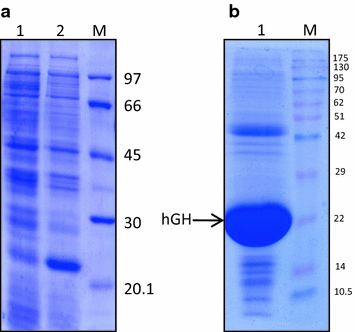


Recovery Of Bioactive Protein From Bacterial Inclusion Bodies Using Trifluoroethanol As Solubilization Agent Microbial Cell Factories Full Text



Guanidinium Chloride Wikipedia



Lessons From Pressure Denaturation Of Proteins Journal Of The Royal Society Interface
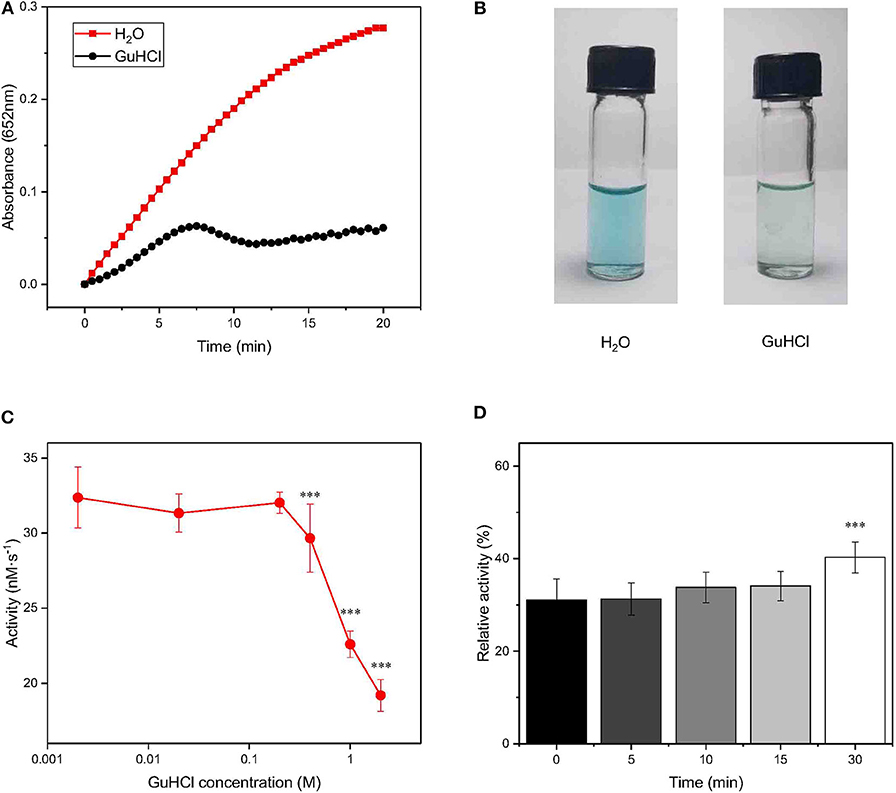


Frontiers Reversible Inhibition Of Iron Oxide Nanozyme By Guanidine Chloride Chemistry



Dynamic Surface Properties Of Lysozyme Solutions Impact Of Urea And Guanidine Hydrochloride Sciencedirect



Denaturation Of Proteins In Guanidine Hcl Fluorescence Of 0 3 M Download Scientific Diagram



Urea But Not Guanidinium Destabilizes Proteins By Forming Hydrogen Bonds To The Peptide Group Pnas



Kinetic Evidence For A Two Stage Mechanism Of Protein Denaturation By Guanidinium Chloride Pnas
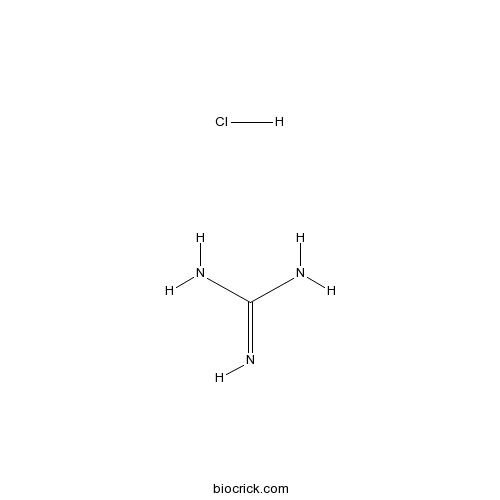


Guanidine Hcl Cas 50 01 1 High Purity Manufacturer Biocrick



Destruction And Building Of The Secondary And Tertiary



Table 1 From Interactions Between Hydrophobic And Ionic Solutes In Aqueous Guanidinium Chloride And Urea Solutions Lessons For Protein Denaturation Mechanism Semantic Scholar



Pdf Denaturation Of Proteins By Urea And Guanidine Hydrochloride


Denaturation Biochemistry Wikipedia



Preparation And Extraction Of Insoluble Inclusion Body Proteins From Escherichia Coli Palmer 04 Current Protocols In Protein Science Wiley Online Library


Investigation Of The Mechanism Of Protein Denaturation By Guanidine Hydrochloride On Vimeo


Protein Denaturation



Guanidinium Hcl Also Used To Disrupt H Bonds In Protein Denaturation Biochemistry Chemistry Bond



Kinetic Evidence For A Two Stage Mechanism Of Protein Denaturation By Guanidinium Chloride Pnas



Kinetic Evidence For A Two Stage Mechanism Of Protein Denaturation By Guanidinium Chloride Pnas



Guanidine Hcl Zellbio Gmbh Quality And Affordable Reagents
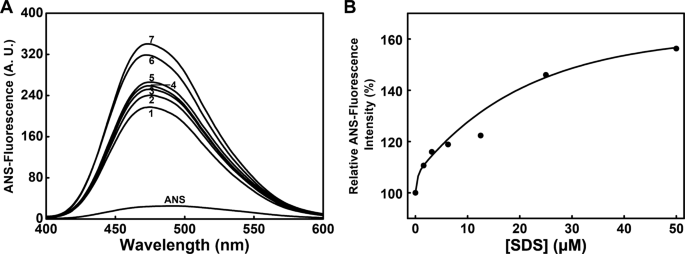


Effects Of Sds On The Activity And Conformation Of Protein Tyrosine Phosphatase From Thermus Thermophilus Hb27 Scientific Reports



Entrapping Intermediates Of Thermal Aggregation In A Helical Proteins With Low Concentration Of Guanidine Hydrochloride Journal Of Biological Chemistry
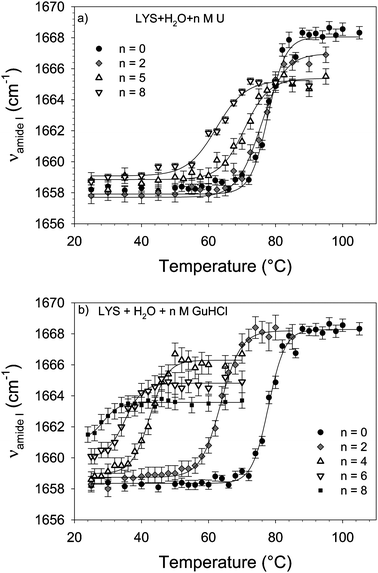


Influence Of Urea And Guanidine Hydrochloride On Lysozyme Stability And Thermal Denaturation A Correlation Between Activity Protein Dynamics And Conformational Changes Physical Chemistry Chemical Physics Rsc Publishing



Comparison Of The Guanidine Hydrochloride Denaturation Curves Of Four Download Scientific Diagram
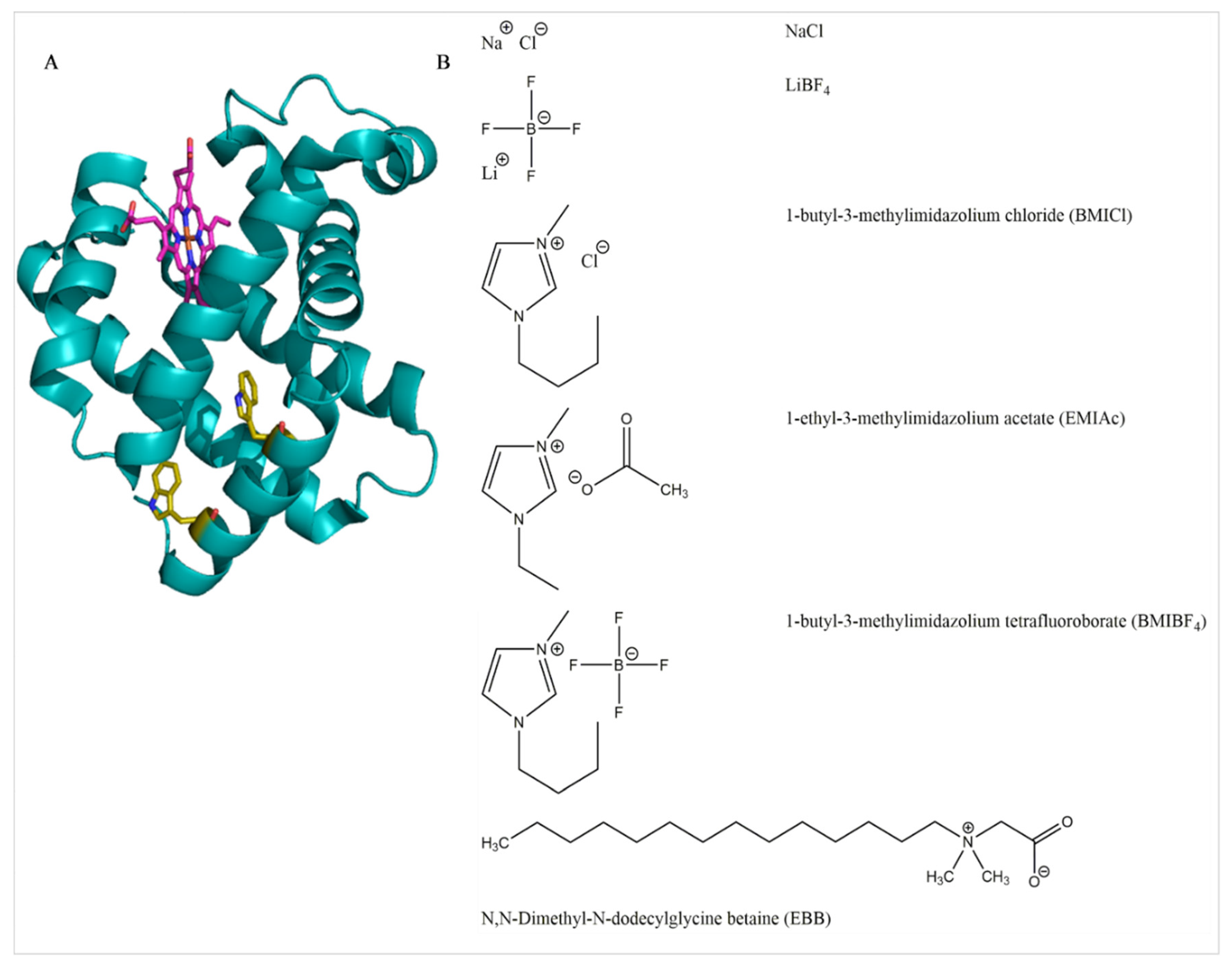


Biomolecules Free Full Text Heme Dissociation From Myoglobin In The Presence Of The Zwitterionic Detergent N N Dimethyl N Dodecylglycine Betaine Effects Of Ionic Liquids Html



Figure 1 From Interactions Between Hydrophobic And Ionic Solutes In Aqueous Guanidinium Chloride And Urea Solutions Lessons For Protein Denaturation Mechanism Semantic Scholar
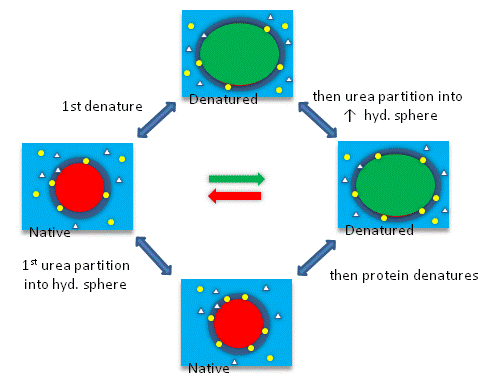


Add Your Page Title
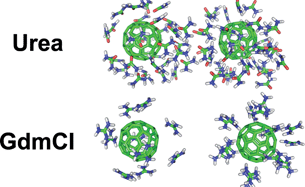


Dissociation Of Hydrophobic And Charged Nano Particles In Aqueous Guanidinium Chloride And Urea Solutions A Molecular Dynamics Study Nanoscale Rsc Publishing



Fluorescence Emission Spectra Of Denatured De Protein A Shift In The Download Scientific Diagram
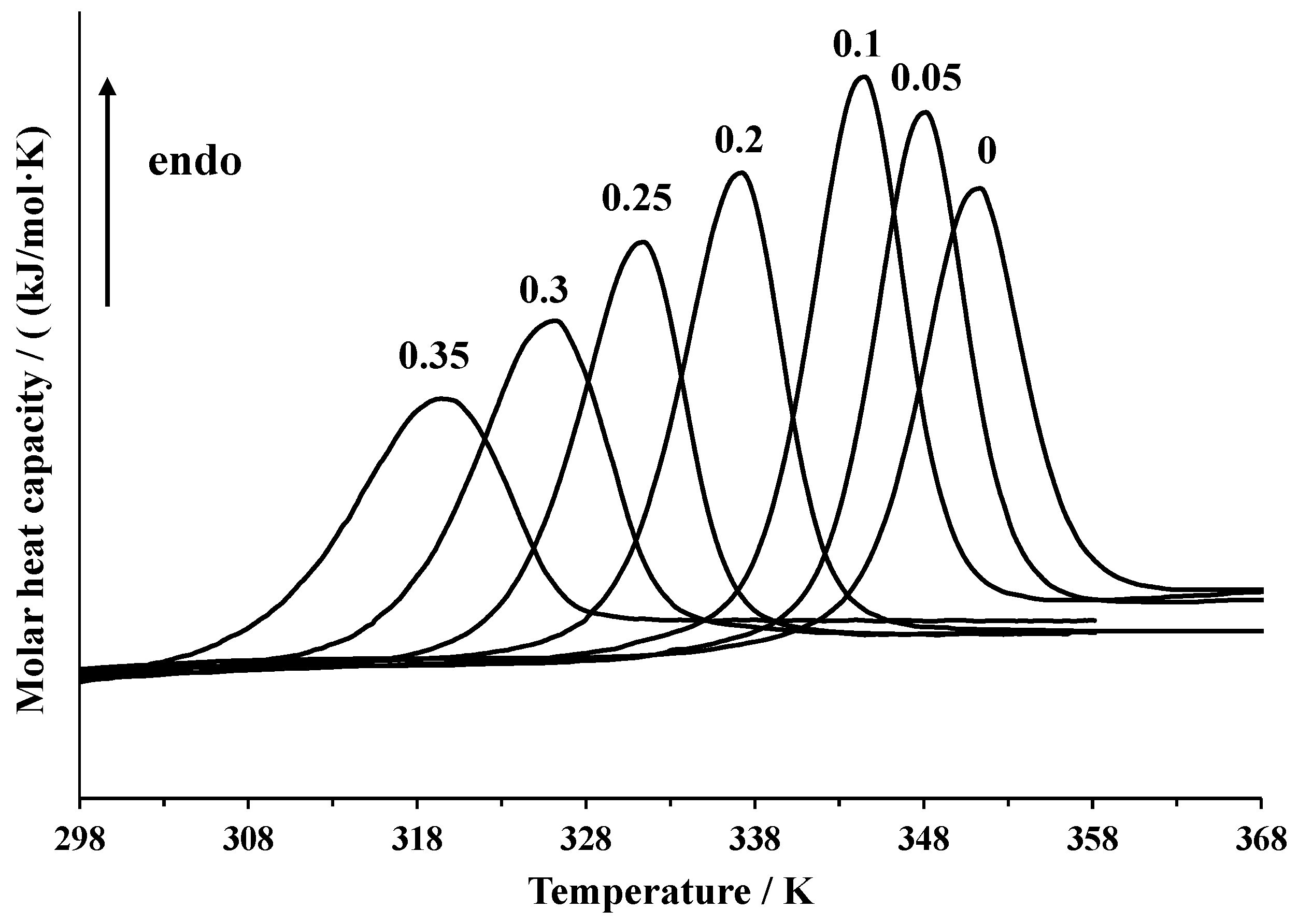


Biomolecules Free Full Text The Effect Of Dimethyl Sulfoxide On The Lysozyme Unfolding Kinetics Thermodynamics And Mechanism Html



Guanidinium Thiocyanate Wikipedia



Metabolism Of Free Guanidine In Bacteria Is Regulated By A Widespread Riboswitch Class Sciencedirect



Interactions Between Hydrophobic And Ionic Solutes In Aqueous Guanidinium Chloride And Urea Solutions Lessons For Protein


Inactivation And Unfolding Of Protein Tyrosine Phosphatase From Thermus Thermophilus Hb27 During Urea And Guanidine Hydrochloride Denaturation


Plos One Inactivation And Unfolding Of Protein Tyrosine Phosphatase From Thermus Thermophilus Hb27 During Urea And Guanidine Hydrochloride Denaturation



Protein Denaturing Agents Youtube
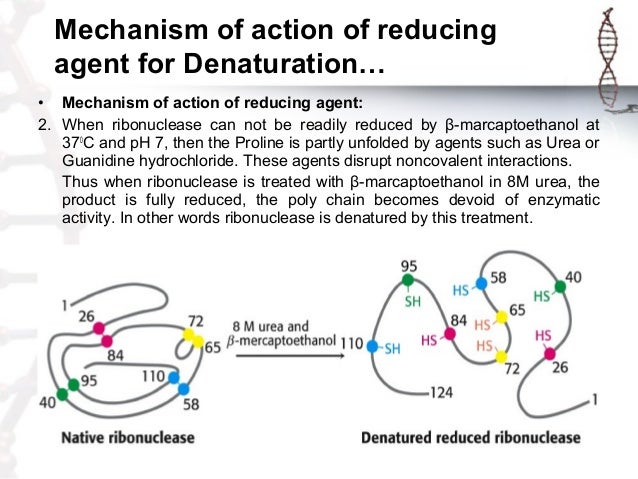


Protein
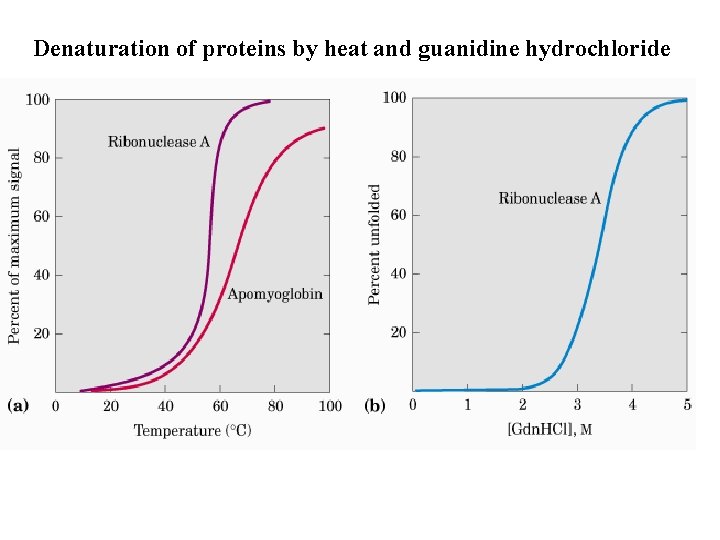


Destruction And Building Of The Secondary And Tertiary
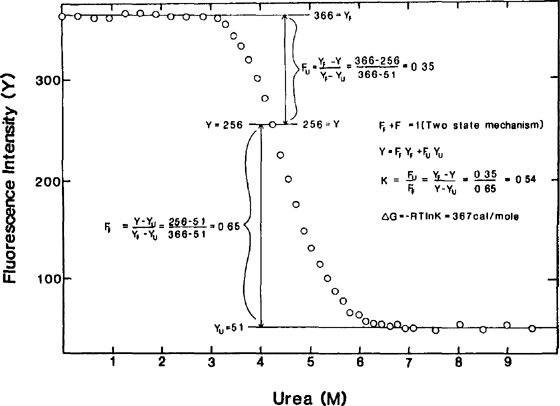


Urea And Guanidine Hydrochloride Denaturation Curves Springerlink


Cooperative Unfolding Of Residual Structure In Heat Denatured Proteins By Urea And Guanidinium Chloride
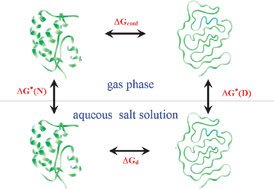


Contrasting The Denaturing Effect Of Guanidinium Chloride With The Stabilizing Effect Of Guanidinium Sulfate Physical Chemistry Chemical Physics Rsc Publishing



Urea And Guanidinium Chloride Denature Protein L In Different Ways In Molecular Dynamics Simulations Biophysical Journal
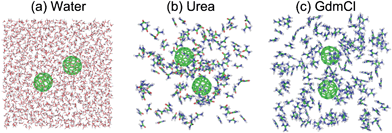


Dissociation Of Hydrophobic And Charged Nano Particles In Aqueous Guanidinium Chloride And Urea Solutions A Molecular Dynamics Study Nanoscale Rsc Publishing
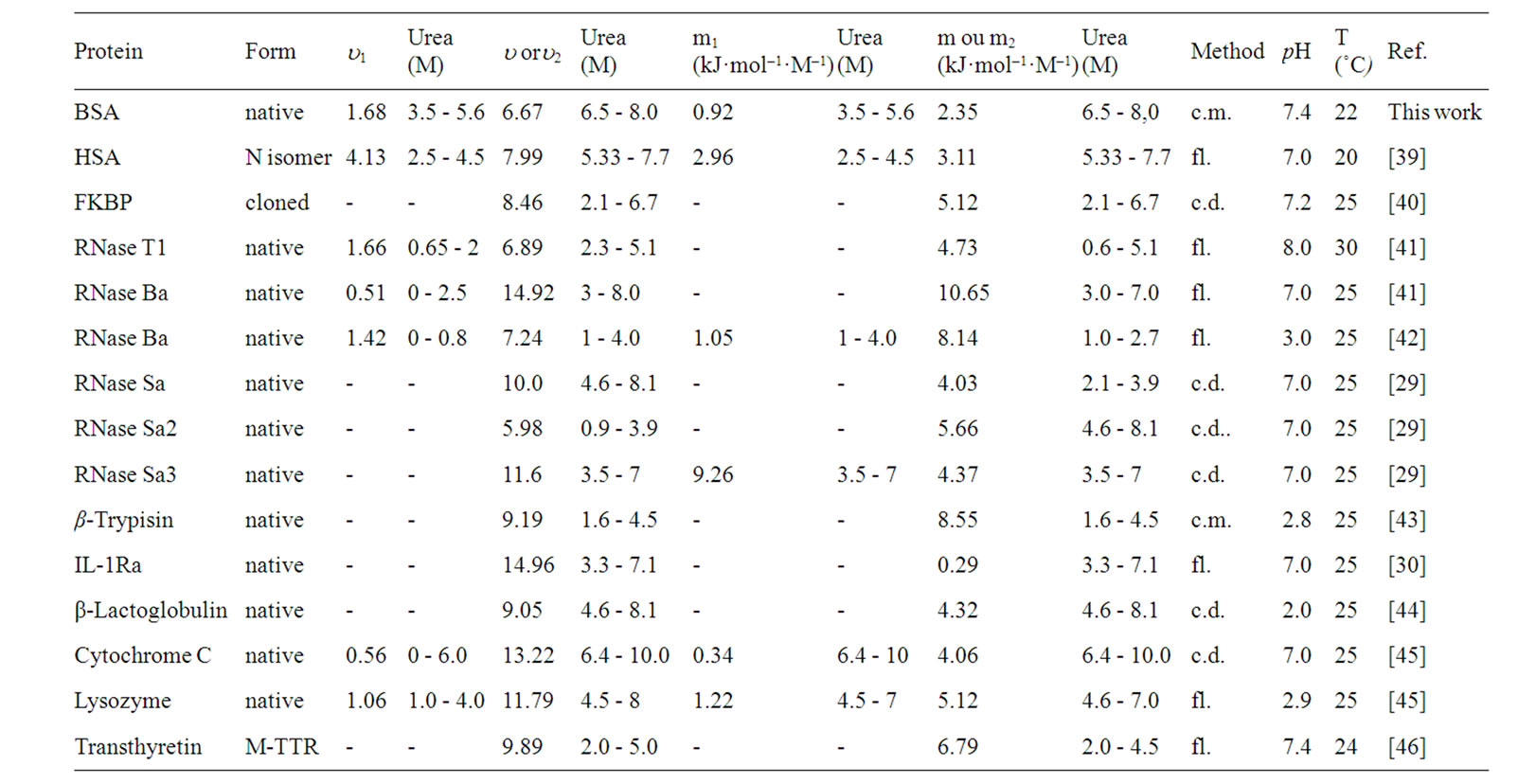


Pressure And Urea Induced Denaturation Of Bovine Serum Albumin Considerations About Protein Heterogeneity
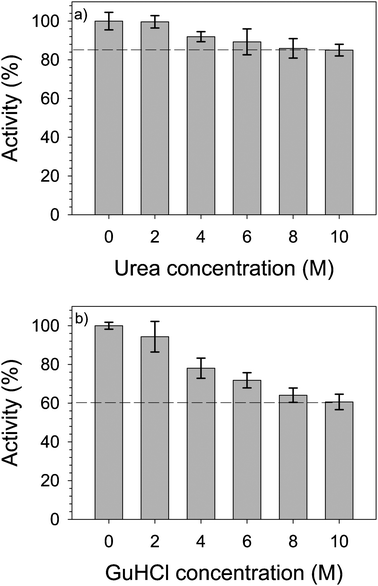


Influence Of Urea And Guanidine Hydrochloride On Lysozyme Stability And Thermal Denaturation A Correlation Between Activity Protein Dynamics And Conformational Changes Physical Chemistry Chemical Physics Rsc Publishing


D Protein Folding Stability In Vivo And In Vitro



0 件のコメント:
コメントを投稿